Sayan Mukherjee, Phillip Febbo, Mauro Maggioni, Jen-Tsan Chi, and Anil Potti.
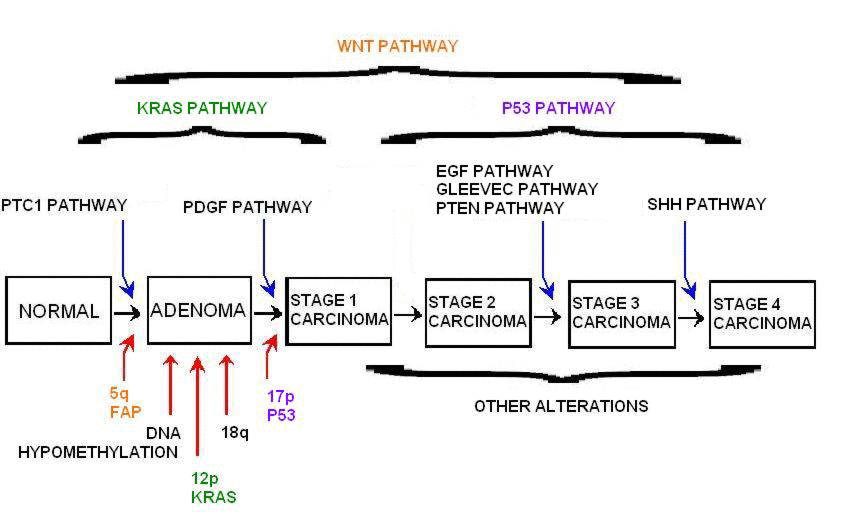
An example of this is developing connections between probability, algebraic topology, and geometry to model complicated dependencies in high dimensional data. The other point of interdisciplinary contact is richer and more complete models in cancer and systems biology as well as population genetics. The modeling approaches being developed will have significant impact in novel patient personalized treatment
strategies.
Ezra Miller, Megan Owen, and Scott Provan
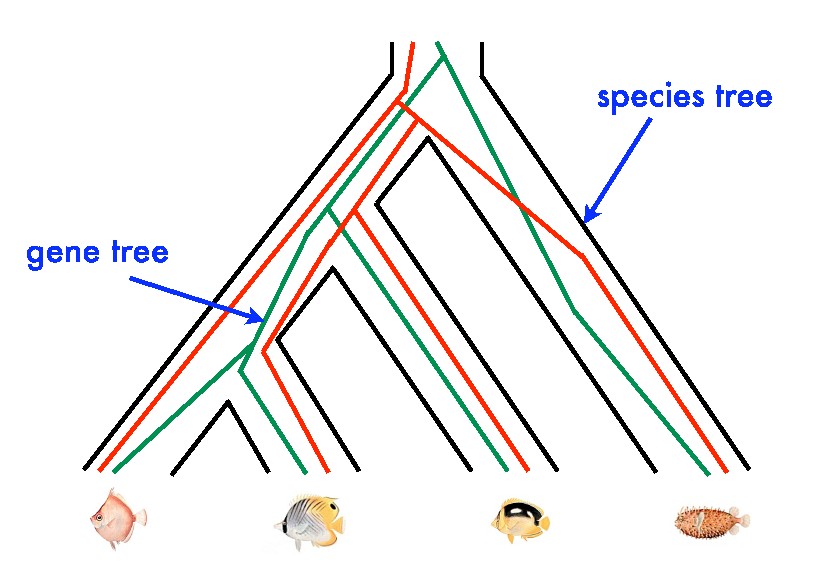
In work with Megan Owen (SAMSI and NC State, Math) and Scott Provan (UNC, Operations Research), we are testing this hypothesis rigorously by developing algorithms for statistical analyses, including computation of statistical means ("centroids") in spaces of phylogenetic trees. These spaces are composed of convex polyhedra -- they are not manifolds, even topologically -- but nonetheless a law of large numbers holds. On the biological side, we will need to find (or construct) and analyze real phylogenetic data sets; this will involve connections with the biology department.
This project has another motivation: in medical imaging, the trees represent branching arteries in brains, and the goal is to detect clustering. This is a special case of analysis of object data, where the general desire is to fit analogues of linear subspaces to data sets whose points represent objects with internal structure, such as curves or trees.